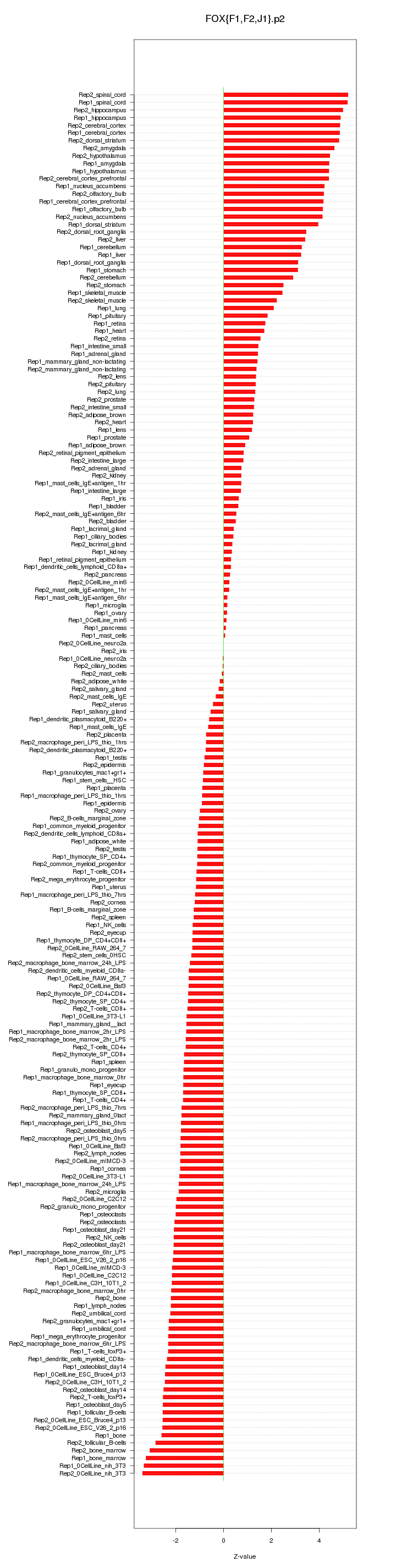
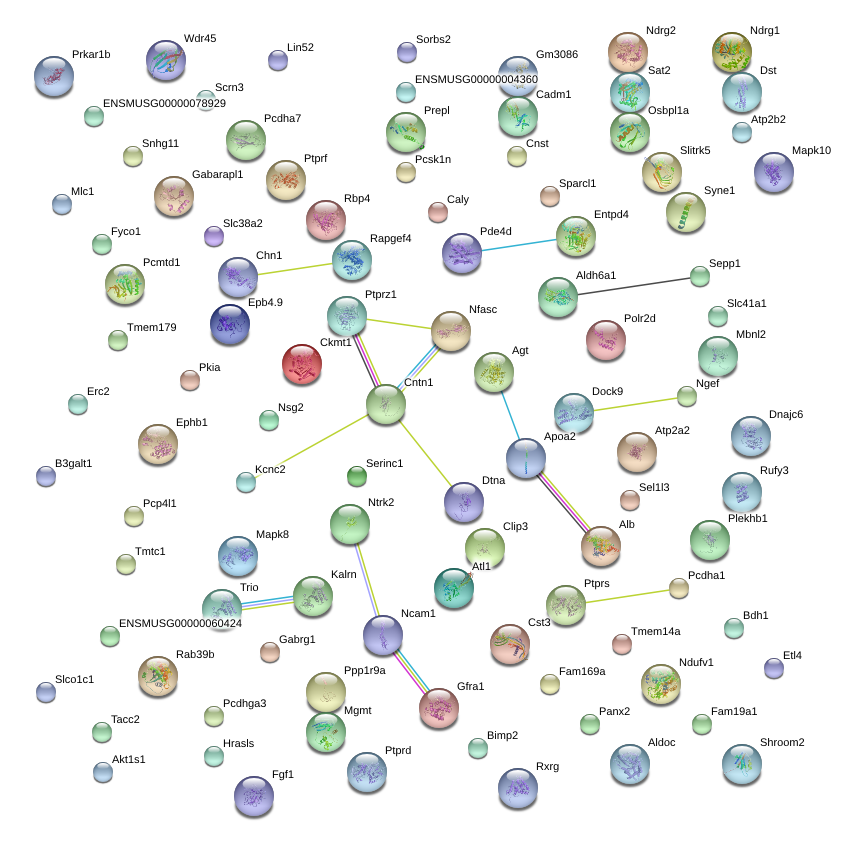
|
chr5_-_104542598
|
113.295
|
|
Sparcl1
|
SPARC-like 1
|
|
chr5_-_104542751
|
91.210
|
|
Sparcl1
|
SPARC-like 1
|
|
chr5_-_104542729
|
90.794
|
|
Sparcl1
|
SPARC-like 1
|
|
chr5_-_104542641
|
88.504
|
|
Sparcl1
|
SPARC-like 1
|
|
chr15_+_91958558
|
74.364
|
NM_007727
|
Cntn1
|
contactin 1
|
|
chr16_-_34263047
|
61.502
|
|
Kalrn
|
kalirin, RhoGEF kinase
|
|
chr16_-_34263272
|
59.566
|
|
Kalrn
|
kalirin, RhoGEF kinase
|
|
chr14_-_52533114
|
56.817
|
NM_001145959
NM_013864
|
Ndrg2
|
N-myc downstream regulated gene 2
|
|
chr14_-_52533039
|
56.605
|
|
Ndrg2
|
N-myc downstream regulated gene 2
|
|
chr14_-_52532988
|
53.878
|
|
Ndrg2
|
N-myc downstream regulated gene 2
|
|
chr1_-_89470442
|
45.015
|
NM_001111314
|
Ngef
|
neuronal guanine nucleotide exchange factor
|
|
chr5_+_89012596
|
41.641
|
NM_027530
|
Rufy3
|
RUN and FYVE domain containing 3
|
|
chr12_-_85791928
|
37.865
|
|
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1
|
|
chr17_-_85461840
|
37.247
|
|
Prepl
|
prolyl endopeptidase-like
|
|
chr7_-_107805365
|
37.235
|
|
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr5_+_89012537
|
37.007
|
|
Rufy3
|
RUN and FYVE domain containing 3
|
|
chr15_+_91991704
|
35.451
|
NM_001159647
|
Cntn1
|
contactin 1
|
|
chr2_+_71892681
|
33.985
|
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr14_+_28689289
|
33.481
|
|
Erc2
|
ELKS/RAB6-interacting/CAST family member 2
|
|
chr12_-_85791897
|
33.281
|
NM_134042
|
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1
|
|
chr7_+_31076771
|
31.559
|
NM_001081114
|
Clip3
|
CAP-GLY domain containing linker protein 3
|
|
chr18_-_39078304
|
31.524
|
NM_010197
|
Fgf1
|
fibroblast growth factor 1
|
|
chr16_+_5885447
|
30.909
|
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr5_+_89012547
|
30.492
|
|
Rufy3
|
RUN and FYVE domain containing 3
|
|
chr15_-_88808967
|
30.225
|
NM_133241
|
Mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 homolog (human)
|
|
chr4_+_101180625
|
29.786
|
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr2_+_20211539
|
29.099
|
NM_001081006
|
Etl4
|
enhancer trap locus 4
|
|
chr3_+_7366586
|
27.616
|
NM_008862
|
Pkia
|
protein kinase inhibitor, alpha
|
|
chr7_+_31076852
|
27.255
|
|
Clip3
|
CAP-GLY domain containing linker protein 3
|
|
chr13_+_59234737
|
26.264
|
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr14_-_71029960
|
26.161
|
NM_013514
|
Epb4.9
|
erythrocyte protein band 4.9
|
|
chr15_+_3220766
|
25.875
|
NM_001042613
NM_001042614
NM_009155
|
Sepp1
|
selenoprotein P, plasma, 1
|
|
chr2_+_73150702
|
24.570
|
NM_029022
|
Scrn3
|
secernin 3
|
|
chr5_-_53499398
|
24.520
|
|
Sel1l3
|
sel-1 suppressor of lin-12-like 3 (C. elegans)
|
|
chr4_-_117881591
|
24.366
|
|
Ptprf
|
protein tyrosine phosphatase, receptor type, F
|
|
chr8_+_46744023
|
24.198
|
|
Sorbs2
|
sorbin and SH3 domain containing 2
|
|
chr2_+_121184334
|
23.966
|
NM_009897
|
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous
|
|
chr5_-_71233855
|
23.530
|
NM_010252
|
Gabrg1
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1
|
|
chr12_+_71008808
|
23.510
|
|
Atl1
|
atlastin GTPase 1
|
|
chr18_+_37089837
|
23.453
|
NM_054072
|
Pcdha1
|
protocadherin alpha 1
|
|
chrX_-_149078963
|
23.345
|
|
Shroom2
|
shroom family member 2
|
|
chr1_+_34068669
|
23.184
|
NM_133833
NM_134448
|
Dst
|
dystonin
|
|
chr18_+_37152024
|
22.888
|
NM_201243
|
Pcdha8
|
protocadherin alpha 8
|
|
chr15_+_3220994
|
22.710
|
|
Sepp1
|
selenoprotein P, plasma, 1
|
|
chr1_+_7162595
|
22.640
|
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1
|
|
chr1_+_34356614
|
22.355
|
|
Dst
|
dystonin
|
|
chr10_+_111708176
|
21.637
|
NM_001025581
|
Kcnc2
|
potassium voltage gated channel, Shaw-related subfamily, member 2
|
|
chr5_-_104543106
|
21.631
|
NM_010097
|
Sparcl1
|
SPARC-like 1
|
|
chrX_+_7496899
|
21.426
|
NM_013892
|
Pcsk1n
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
|
chr19_-_38199737
|
21.420
|
NM_001159487
|
Rbp4
|
retinol binding protein 4, plasma
|
|
chr5_-_139606339
|
20.478
|
|
Prkar1b
|
protein kinase, cAMP dependent regulatory, type I beta
|
|
chr15_+_3220999
|
20.160
|
|
Sepp1
|
selenoprotein P, plasma, 1
|
|
chr5_+_90890764
|
19.560
|
|
Alb
|
albumin
|
|
chr5_-_122906470
|
19.437
|
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chr18_+_37133455
|
19.335
|
NM_009957
|
Pcdha7
|
protocadherin alpha 7
|
|
chr12_-_113749363
|
18.979
|
NM_178915
|
Tmem179
|
transmembrane protein 179
|
|
chr10_+_5151833
|
18.683
|
NM_001079686
|
Syne1
|
synaptic nuclear envelope 1
|
|
chr1_+_9591492
|
18.418
|
|
3110035E14Rik
|
RIKEN cDNA 3110035E14 gene
|
|
chr7_+_137720997
|
18.316
|
NM_001004468
NM_206856
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr3_+_62142698
|
18.130
|
NM_001081295
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26
|
|
chr13_+_97837240
|
17.946
|
NM_001146045
|
Fam169a
|
family with sequence similarity 169, member A
|
|
chr1_+_21208679
|
17.712
|
NM_029398
|
Tmem14a
|
transmembrane protein 14A
|
|
chr19_-_58529766
|
17.656
|
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1
|
|
chr9_+_47338431
|
17.388
|
NM_001025600
NM_018770
NM_207675
NM_207676
|
Cadm1
|
cell adhesion molecule 1
|
|
chr18_+_23468655
|
17.388
|
|
Dtna
|
dystrobrevin alpha
|
|
chrX_-_72823561
|
17.337
|
|
Rab39b
|
RAB39B, member RAS oncogene family
|
|
chr9_+_47338255
|
17.150
|
|
Cadm1
|
cell adhesion molecule 1
|
|
chrX_+_7299358
|
16.877
|
NM_172372
|
Wdr45
|
WD repeat domain 45
|
|
chr6_-_148392841
|
16.817
|
NM_198967
|
Tmtc1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr6_+_129483238
|
16.755
|
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1
|
|
chr13_+_109444370
|
16.714
|
NM_011056
|
Pde4d
|
phosphodiesterase 4D, cAMP specific
|
|
chr6_+_129483295
|
16.668
|
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1
|
|
chr2_-_73498303
|
16.461
|
NM_001166604
NM_175752
|
Chn1
|
chimerin (chimaerin) 1
|
|
chr4_+_101180580
|
16.244
|
NM_001164584
NM_001164585
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr9_-_49311823
|
16.227
|
|
Ncam1
|
neural cell adhesion molecule 1
|
|
chr1_-_134603848
|
15.948
|
NM_001160318
|
Nfasc
|
neurofascin
|
|
chr16_+_29209780
|
15.573
|
NM_013751
|
Hrasls
|
HRAS-like suppressor
|
|
chr1_-_173126368
|
15.566
|
NM_025557
|
Pcp4l1
|
Purkinje cell protein 4-like 1
|
|
chr7_-_107805466
|
15.562
|
NM_001163186
NM_001163187
|
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chrX_+_7299332
|
15.447
|
|
Wdr45
|
WD repeat domain 45
|
|
chr15_-_66801093
|
15.396
|
|
Ndrg1
|
N-myc downstream regulated gene 1
|
|
chr12_+_85792371
|
15.204
|
NM_173756
|
Lin52
|
lin-52 homolog (C. elegans)
|
|
chr11_+_78137699
|
14.948
|
NM_009657
|
Aldoc
|
aldolase C, fructose-bisphosphate
|
|
chr19_-_4012698
|
14.923
|
NM_133666
|
Ndufv1
|
NADH dehydrogenase (ubiquinone) flavoprotein 1
|
|
chr19_-_4012654
|
14.862
|
|
Ndufv1
|
NADH dehydrogenase (ubiquinone) flavoprotein 1
|
|
chr1_+_173155414
|
14.849
|
|
Apoa2
|
apolipoprotein A-II
|
|
chr14_+_120674921
|
14.636
|
|
Mbnl2
|
muscleblind-like 2
|
|
chr11_+_69435553
|
14.527
|
NM_026991
|
Sat2
|
spermidine/spermine N1-acetyl transferase 2
|
|
chr18_-_13021181
|
14.508
|
|
Osbpl1a
|
oxysterol binding protein-like 1A
|
|
chr14_+_120674939
|
14.488
|
|
Mbnl2
|
muscleblind-like 2
|
|
chr18_+_23468585
|
14.374
|
|
Dtna
|
dystrobrevin alpha
|
|
chr14_+_120674933
|
14.351
|
|
Mbnl2
|
muscleblind-like 2
|
|
chr10_+_28931448
|
14.349
|
NM_001162537
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene
|
|
chr6_+_4853319
|
14.218
|
NM_181595
|
Ppp1r9a
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr6_+_96063137
|
14.167
|
NM_182808
|
Fam19a1
|
family with sequence similarity 19, member A1
|
|
chr2_+_67955769
|
14.155
|
NM_020283
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1
|
|
chr2_+_158201373
|
14.081
|
NM_175692
|
Snhg11
|
small nucleolar RNA host gene 11
|
|
chr9_-_102256962
|
13.949
|
NM_001168296
NM_173447
|
Ephb1
|
Eph receptor B1
|
|
chr14_+_112074336
|
13.907
|
NM_198865
|
Slitrk5
|
SLIT and NTRK-like family, member 5
|
|
chr5_-_103640272
|
13.886
|
NM_001081567
NM_009158
|
Mapk10
|
mitogen-activated protein kinase 10
|
|
chr18_+_37854011
|
13.857
|
NM_033588
|
Pcdhga5
|
protocadherin gamma subfamily A, 5
|
|
chr10_-_57252251
|
13.690
|
|
Serinc1
|
serine incorporator 1
|
|
chr18_+_37112301
|
13.681
|
NM_001174154
NM_007766
|
Pcdha4-g
Pcdha4
|
protocadherin alpha 4-gamma
protocadherin alpha 4
|
|
chr7_-_147268395
|
13.655
|
NM_001190385
NM_026769
|
Caly
|
calcyon neuron-specific vesicular protein
|
|
chr6_+_141472906
|
13.345
|
NM_001177772
NM_021471
|
Slco1c1
|
solute carrier organic anion transporter family, member 1c1
|
|
chr1_+_181476534
|
13.119
|
NM_146105
|
Cnst
|
consortin, connexin sorting protein
|
|
chr14_-_122097583
|
13.056
|
NM_001081039
|
Dock9
|
dedicator of cytokinesis 9
|
|
chr1_-_42749940
|
12.924
|
|
2610017I09Rik
|
RIKEN cDNA 2610017I09 gene
|
|
chr16_+_31422377
|
12.838
|
NM_175177
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1
|
|
chr12_+_110834872
|
12.751
|
|
1110006E14Rik
|
RIKEN cDNA 1110006E14 gene
|
|
chr10_+_122701051
|
12.675
|
NM_182807
|
Fam19a2
|
family with sequence similarity 19, member A2
|
|
chr1_+_133737686
|
12.584
|
|
Slc41a1
|
solute carrier family 41, member 1
|
|
chr14_+_69984344
|
12.480
|
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4
|
|
chr14_+_120674913
|
12.457
|
|
Mbnl2
|
muscleblind-like 2
|
|
chr2_-_148701164
|
12.408
|
|
Cst3
|
cystatin C
|
|
chr15_-_66801198
|
12.367
|
NM_008681
|
Ndrg1
|
N-myc downstream regulated gene 1
|
|
chr1_+_169548381
|
12.344
|
NM_001159731
|
Rxrg
|
retinoid X receptor gamma
|
|
chr5_+_89012600
|
12.301
|
|
Rufy3
|
RUN and FYVE domain containing 3
|
|
chr15_+_88890519
|
12.050
|
|
Panx2
|
pannexin 2
|
|
chr7_+_52104591
|
11.944
|
NM_026270
|
Akt1s1
|
AKT1 substrate 1 (proline-rich)
|
|
chr6_-_113841356
|
11.908
|
NM_001036684
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr8_-_127093601
|
11.871
|
NM_007428
|
Agt
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr6_+_129483215
|
11.852
|
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1
|
|
chr6_+_22825464
|
11.837
|
NM_001081306
|
Ptprz1
|
protein tyrosine phosphatase, receptor type Z, polypeptide 1
|
|
chr11_+_31900378
|
11.787
|
NM_008741
|
Nsg2
|
neuron specific gene family member 2
|
|
chr15_+_99223377
|
11.786
|
NM_001171034
NM_001171036
NM_026669
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6
|
|
chr4_-_118162354
|
11.773
|
|
Tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1
|
|
chr2_-_127481868
|
11.717
|
|
Mal
|
myelin and lymphocyte protein, T-cell differentiation protein
|
|
chr18_-_12978106
|
11.601
|
|
Osbpl1a
|
oxysterol binding protein-like 1A
|
|
chr4_-_118162385
|
11.583
|
NM_011587
|
Tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1
|
|
chr4_-_62815137
|
11.543
|
NM_007443
|
Ambp
|
alpha 1 microglobulin/bikunin
|
|
chr2_+_26828935
|
11.386
|
NM_001001322
|
Adamts13
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 13
|
|
chr1_+_9591550
|
11.249
|
|
3110035E14Rik
|
RIKEN cDNA 3110035E14 gene
|
|
chr6_+_129483154
|
11.180
|
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1
|
|
chr9_-_107571294
|
11.159
|
|
Slc38a3
|
solute carrier family 38, member 3
|
|
chr13_-_111070225
|
11.052
|
|
Rab3c
|
RAB3C, member RAS oncogene family
|
|
chr10_+_94038001
|
10.985
|
NM_001168684
|
Tmcc3
|
transmembrane and coiled coil domains 3
|
|
chr3_-_97692613
|
10.950
|
NM_001039376
|
Pde4dip
|
phosphodiesterase 4D interacting protein (myomegalin)
|
|
chr6_-_60779848
|
10.950
|
NM_009221
|
Snca
|
synuclein, alpha
|
|
chr16_+_42955690
|
10.934
|
|
BC002163
|
NADH dehydrogenase Fe-S protein 5 pseudogene
|
|
chr2_-_71205805
|
10.927
|
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12
|
|
chr14_-_104243541
|
10.795
|
|
Ednrb
|
endothelin receptor type B
|
|
chr8_+_73051625
|
10.713
|
NM_001111066
NM_010223
|
Fkbp8
|
FK506 binding protein 8
|
|
chr7_-_125263314
|
10.653
|
|
Arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1
|
|
chr15_-_63891810
|
10.597
|
NM_144846
|
Fam49b
|
family with sequence similarity 49, member B
|
|
chr7_+_148817700
|
10.536
|
|
Ap2a2
|
adaptor protein complex AP-2, alpha 2 subunit
|
|
chr10_-_57252252
|
10.399
|
NM_019760
|
Serinc1
|
serine incorporator 1
|
|
chr15_+_97942484
|
10.355
|
NM_021514
|
Pfkm
|
phosphofructokinase, muscle
|
|
chr6_+_49773596
|
10.275
|
|
Npy
|
neuropeptide Y
|
|
chr3_+_105916283
|
10.214
|
NM_023186
|
Chia
|
chitinase, acidic
|
|
chr9_-_107571304
|
10.201
|
|
Slc38a3
|
solute carrier family 38, member 3
|
|
chr8_-_87297283
|
10.200
|
|
Nfix
|
nuclear factor I/X
|
|
chr7_-_93924317
|
10.171
|
NM_001159706
NM_016770
|
Folh1
|
folate hydrolase
|
|
chr19_+_53403902
|
10.100
|
NM_010847
|
Mxi1
|
Max interacting protein 1
|
|
chr11_-_97372344
|
10.045
|
|
Srcin1
|
SRC kinase signaling inhibitor 1
|
|
chr7_-_26460180
|
9.971
|
NM_177102
|
Tmem91
|
transmembrane protein 91
|
|
chr2_-_91019477
|
9.915
|
NM_001177720
|
Madd
|
MAP-kinase activating death domain
|
|
chr9_-_73781373
|
9.897
|
NM_001081153
|
Unc13c
|
unc-13 homolog C (C. elegans)
|
|
chr14_-_94287950
|
9.887
|
NM_001081377
|
Pcdh9
|
protocadherin 9
|
|
chr19_-_29101759
|
9.880
|
|
|
|
|
chr1_-_182126032
|
9.774
|
NM_023341
|
Adck3
|
aarF domain containing kinase 3
|
|
chr8_+_73051648
|
9.725
|
|
Fkbp8
|
FK506 binding protein 8
|
|
chr12_+_105576693
|
9.674
|
NM_011458
|
Serpina3k
|
serine (or cysteine) peptidase inhibitor, clade A, member 3K
|
|
chr6_+_129483206
|
9.670
|
NM_020590
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1
|
|
chr5_-_113709569
|
9.636
|
|
Sgsm1
|
small G protein signaling modulator 1
|
|
chr8_+_73051651
|
9.580
|
|
Fkbp8
|
FK506 binding protein 8
|
|
chr11_-_107209443
|
9.567
|
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1
|
|
chr17_+_75835208
|
9.435
|
NM_001166493
|
Rasgrp3
|
RAS, guanyl releasing protein 3
|
|
chr9_+_47338449
|
9.373
|
|
Cadm1
|
cell adhesion molecule 1
|
|
chr6_-_113841295
|
9.254
|
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr12_+_105644917
|
9.216
|
NM_009252
|
Serpina3n
|
serine (or cysteine) peptidase inhibitor, clade A, member 3N
|
|
chr9_+_46036788
|
9.204
|
|
Apoa1
|
apolipoprotein A-I
|
|
chr7_+_107423813
|
9.171
|
|
Pgm2l1
|
phosphoglucomutase 2-like 1
|
|
chr15_+_99223622
|
9.069
|
NM_001171035
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6
|
|
chr7_-_26460133
|
9.066
|
|
Tmem91
|
transmembrane protein 91
|
|
chr18_+_37844927
|
9.040
|
NM_033587
|
Pcdhga4
|
protocadherin gamma subfamily A, 4
|
|
chr1_-_94538517
|
8.984
|
NM_001163425
|
Myeov2
|
myeloma overexpressed 2
|
|
chr19_-_42160732
|
8.973
|
NM_198108
|
Morn4
|
MORN repeat containing 4
|
|
chr11_-_103733519
|
8.830
|
|
Nsf
|
N-ethylmaleimide sensitive fusion protein
|
|
chr17_+_24546733
|
8.733
|
|
Abca3
|
ATP-binding cassette, sub-family A (ABC1), member 3
|
|
chr3_-_146432922
|
8.713
|
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta
|
|
chr7_-_52104247
|
8.646
|
|
Tbc1d17
|
TBC1 domain family, member 17
|
|
chr5_-_138181225
|
8.595
|
NM_175521
|
6430598A04Rik
|
RIKEN cDNA 6430598A04 gene
|
|
chr11_+_69867869
|
8.551
|
NM_009714
|
Asgr1
|
asialoglycoprotein receptor 1
|
|
chr2_+_164311755
|
8.547
|
NM_026797
|
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr19_-_21467250
|
8.539
|
|
Gda
|
guanine deaminase
|
|
chr1_+_91351373
|
8.401
|
NM_001037136
NM_178119
|
Agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr3_+_88373199
|
8.349
|
|
Ubqln4
|
ubiquilin 4
|
|
chr12_+_38607291
|
8.298
|
NM_178681
|
Dgkb
|
diacylglycerol kinase, beta
|
|
chr6_+_38484796
|
8.294
|
NM_025363
|
1110001J03Rik
|
RIKEN cDNA 1110001J03 gene
|
|
chr10_-_57252184
|
8.284
|
|
Serinc1
|
serine incorporator 1
|
|
chr1_-_170085798
|
8.230
|
|
Pbx1
|
pre B-cell leukemia transcription factor 1
|
|
chr1_-_174128558
|
8.217
|
|
Pea15a
|
phosphoprotein enriched in astrocytes 15A
|
|
chr8_-_47209499
|
8.045
|
NM_207213
|
Snx25
|
sorting nexin 25
|
|
chr8_+_96334879
|
8.029
|
|
Gnao1
|
guanine nucleotide binding protein, alpha O
|
|
chr2_-_32702729
|
8.027
|
NM_009295
NM_001113569
|
Stxbp1
|
syntaxin binding protein 1
|
|
chr11_+_16157685
|
7.953
|
NM_145967
|
Vstm2a
|
V-set and transmembrane domain containing 2A
|
|
chr16_+_7409879
|
7.927
|
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr7_-_52104448
|
7.814
|
NM_001042655
|
Tbc1d17
|
TBC1 domain family, member 17
|
|
chr9_+_46036799
|
7.772
|
|
Apoa1
|
apolipoprotein A-I
|